-Search query
-Search result
Showing 1 - 50 of 160 items for (author: garcia & ba)

EMDB-16916: 
Bipartite interaction of TOPBP1 with the GINS complex
Method: single particle / : Day M, Oliver AW, Pearl LH

PDB-8ok2: 
Bipartite interaction of TOPBP1 with the GINS complex
Method: single particle / : Day M, Oliver AW, Pearl LH

EMDB-16427: 
Apo Hantaan virus polymerase core
Method: single particle / : Durieux trouilleton Q, Arragain B, Malet H

EMDB-16428: 
Hantaan virus polymerase bound to its 5' viral RNA
Method: single particle / : Durieux trouilleton Q, Arragain B, Malet H

EMDB-16429: 
Hantaan virus polymerase in replication pre-initiation state
Method: single particle / : Durieux trouilleton Q, Arragain B, Malet H

EMDB-16430: 
Hantaan virus polymerase in replication elongation state
Method: single particle / : Durieux trouilleton Q, Arragain B, Malet H

EMDB-28649: 
Structure of Janus Kinase (JAK) dimer complexed with cytokine receptor intracellular domain
Method: single particle / : Caveney NA, Saxton RA, Waghray D, Garcia KC

PDB-8ewy: 
Structure of Janus Kinase (JAK) dimer complexed with cytokine receptor intracellular domain
Method: single particle / : Caveney NA, Saxton RA, Waghray D, Garcia KC

EMDB-14738: 
Cryo-EM structure of hnRNPDL amyloid fibrils
Method: helical / : Chaves-Sanjuan A, Garcia-Pardo J, Bartolome-Nafria A, Gil-Garcia M, Visentin C, Bolognesi M, Ricagno S, Ventura S

PDB-7zir: 
Cryo-EM structure of hnRNPDL amyloid fibrils
Method: helical / : Garcia-Pardo J, Chaves-Sanjuan A, Bartolome-Nafria A, Gil-Garcia M, Visentin C, Bolognesi M, Ricagno S, Ventura S

EMDB-14472: 
Structure of the RAF1-HSP90-CDC37 complex (RHC-II)
Method: single particle / : Mesa P, Garcia-Alonso S, Barbacid M, Montoya G

EMDB-14473: 
Structure of the RAF1-HSP90-CDC37 complex (RHC-I)
Method: single particle / : Mesa P, Garcia-Alonso S, Barbacid M, Montoya G

PDB-7z37: 
Structure of the RAF1-HSP90-CDC37 complex (RHC-II)
Method: single particle / : Mesa P, Garcia-Alonso S, Barbacid M, Montoya G

PDB-7z38: 
Structure of the RAF1-HSP90-CDC37 complex (RHC-I)
Method: single particle / : Mesa P, Garcia-Alonso S, Barbacid M, Montoya G

EMDB-26372: 
SARS-2 CoV 6P Mut7 in complex with Fab CC84.5
Method: single particle / : Torres JL, Ward AB

EMDB-26365: 
SARS-2 CoV 6P Mut7 in complex with Fab CC25.1
Method: single particle / : Torres JL, Ward AB
EMDB-26366: 
SARS-2 CoV 6P Mut7 in complex with Fab CC25.54
Method: single particle / : Torres JL, Ward AB

EMDB-26367: 
SARS-2 CoV 6P Mut7 + Fab CC25.52
Method: single particle / : Torres JL, Ward AB

EMDB-26368: 
SARS-2 CoV 6P Mut7 in complex with Fab CC25.56
Method: single particle / : Torres JL, Ward AB
EMDB-26369: 
SARS-2 CoV 6P Mut7 in complex with Fab CC25.36
Method: single particle / : Torres JL, Ward AB

EMDB-26370: 
SARS-2 CoV 6P Mut7 in complex with Fab CC25.3
Method: single particle / : Torres JL, Ward AB
EMDB-26371: 
SARS-2 CoV 6P Mut7 in complex with Fab CC25.11
Method: single particle / : Torres JL, Ward AB

EMDB-26373: 
SARS-2 CoV 6P Mut7 in complex with Fab CC84.2
Method: single particle / : Torres JL, Ward AB

EMDB-26374: 
SARS-2 CoV 6P Mut7 in complex with Fab CC84.10
Method: single particle / : Torres JL, Ward AB
EMDB-26375: 
SARS-2 CoV 6P Mut7 in complex with Fab CC84.24
Method: single particle / : Torres JL, Ward AB

EMDB-23887: 
Complex structure of trailing EC of EC+EC (trailing EC-focused)
Method: single particle / : Yang C, Murakami K

EMDB-23888: 
Structure of EC+EC (leading EC-focused)
Method: single particle / : Yang C, Murakami K

PDB-7mk9: 
Complex structure of trailing EC of EC+EC (trailing EC-focused)
Method: single particle / : Yang C, Murakami K

PDB-7mka: 
Structure of EC+EC (leading EC-focused)
Method: single particle / : Yang C, Murakami K

EMDB-23789: 
Composite structure of EC+EC
Method: single particle / : Yang C, Murakami K

PDB-7mei: 
Composite structure of EC+EC
Method: single particle / : Yang C, Murakami K

EMDB-23904: 
RNA polymerase II pre-initiation complex (PIC1)
Method: single particle / : Yang C, Fujiwara R, Kim HJ, Gorbea Colon JJ, Steimle S, Garcia BA, Murakami K

EMDB-23905: 
RNA polymerase II pre-initiation complex (PIC2)
Method: single particle / : Yang C, Fujiwara R, Kim HJ, Gorbea Colon JJ, Steimle S, Garcia BA, Murakami K

EMDB-23906: 
RNA polymerase II pre-initiation complex (PIC3)
Method: single particle / : Yang C, Fujiwara R, Kim HJ, Gorbea Colon JJ, Steimle S, Garcia BA, Murakami K

EMDB-23907: 
General transcription factor TFIIH (weak binding)
Method: single particle / : Yang C, Fujiwara R, Kim HJ, Gorbea Colon JJ, Steimle S, Garcia BA, Murakami K

EMDB-23908: 
RNA polymerase II initially transcribing complex (ITC)
Method: single particle / : Yang C, Fujiwara R, Kim HJ, Gorbea Colon JJ, Steimle S, Garcia BA, Murakami K

PDB-7ml0: 
RNA polymerase II pre-initiation complex (PIC1)
Method: single particle / : Yang C, Fujiwara R, Kim HJ, Gorbea Colon JJ, Steimle S, Garcia BA, Murakami K

PDB-7ml1: 
RNA polymerase II pre-initiation complex (PIC2)
Method: single particle / : Yang C, Fujiwara R, Kim HJ, Gorbea Colon JJ, Steimle S, Garcia BA, Murakami K

PDB-7ml2: 
RNA polymerase II pre-initiation complex (PIC3)
Method: single particle / : Yang C, Fujiwara R, Kim HJ, Gorbea Colon JJ, Steimle S, Garcia BA, Murakami K

PDB-7ml3: 
General transcription factor TFIIH (weak binding)
Method: single particle / : Yang C, Fujiwara R, Kim HJ, Gorbea Colon JJ, Steimle S, Garcia BA, Murakami K

PDB-7ml4: 
RNA polymerase II initially transcribing complex (ITC)
Method: single particle / : Yang C, Fujiwara R, Kim HJ, Gorbea Colon JJ, Steimle S, Garcia BA, Murakami K

EMDB-25634: 
Negative stain map of monoclonal Fab 047-09 4F04 binding the anchor epitope of H1 HA
Method: single particle / : Han J, Richey ST, Ward AB

EMDB-25635: 
Negative stain map of monoclonal Fab 241 IgA 2F04 binding the anchor epitope of H1 HA
Method: single particle / : Han J, Richey ST, Ward AB

EMDB-25636: 
Negative stain map of polyclonal Fab 236.7 binding the anchor and esterase epitopes of H1 HA
Method: single particle / : Han J, Richey ST, Ward AB

EMDB-25637: 
Negative stain map of polyclonal Fab 236.7 binding the RBS epitope of H1 HA
Method: single particle / : Han J, Richey ST, Ward AB

EMDB-25638: 
Negative stain map of polyclonal Fab 236.14 binding an epitope on the top of the head of H1 HA
Method: single particle / : Han J, Richey ST, Ward AB
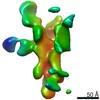
EMDB-25639: 
Negative stain map of polyclonal Fab 236.14 binding the esterase epitope of H1 HA
Method: single particle / : Han J, Richey ST, Ward AB

EMDB-25640: 
Negative stain map of polycolonal Fab 236.14 binding the RBS epitope of H1 HA
Method: single particle / : Han J, Richey ST, Ward AB

EMDB-25641: 
Negative stain map of polyclonal Fab 236.14 binding the anchor epitope of H1 HA
Method: single particle / : Han J, Richey ST, Ward AB
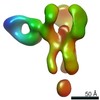
EMDB-25642: 
Negative stain map of polyclonal Fab 241.7 binding the esterase epitope of H1 HA
Method: single particle / : Han J, Richey ST, Ward AB
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
